FTIR Spectroscopy Quality Assurance Tool
A Python desktop application used by lab technicians to assess FTIR spectra quality in real time.
The FTIR Spectroscopy Quality Assurance Tool automates checks on absorbance, signal-to-noise ratio, and water vapour contamination, providing clear pass/fail feedback and visualisations. This has enabled immediate feedback on sample preparation procedures and helped standardise the collection of high-quality samples.
Project description
This project is a standalone quality assurance tool for FTIR (Fourier-transform infrared) spectroscopy data, designed for day-to-day use by lab technicians. The application provides an end-to-end workflow for loading spectral data, running automated quality checks, visualising results, and exporting a structured report.
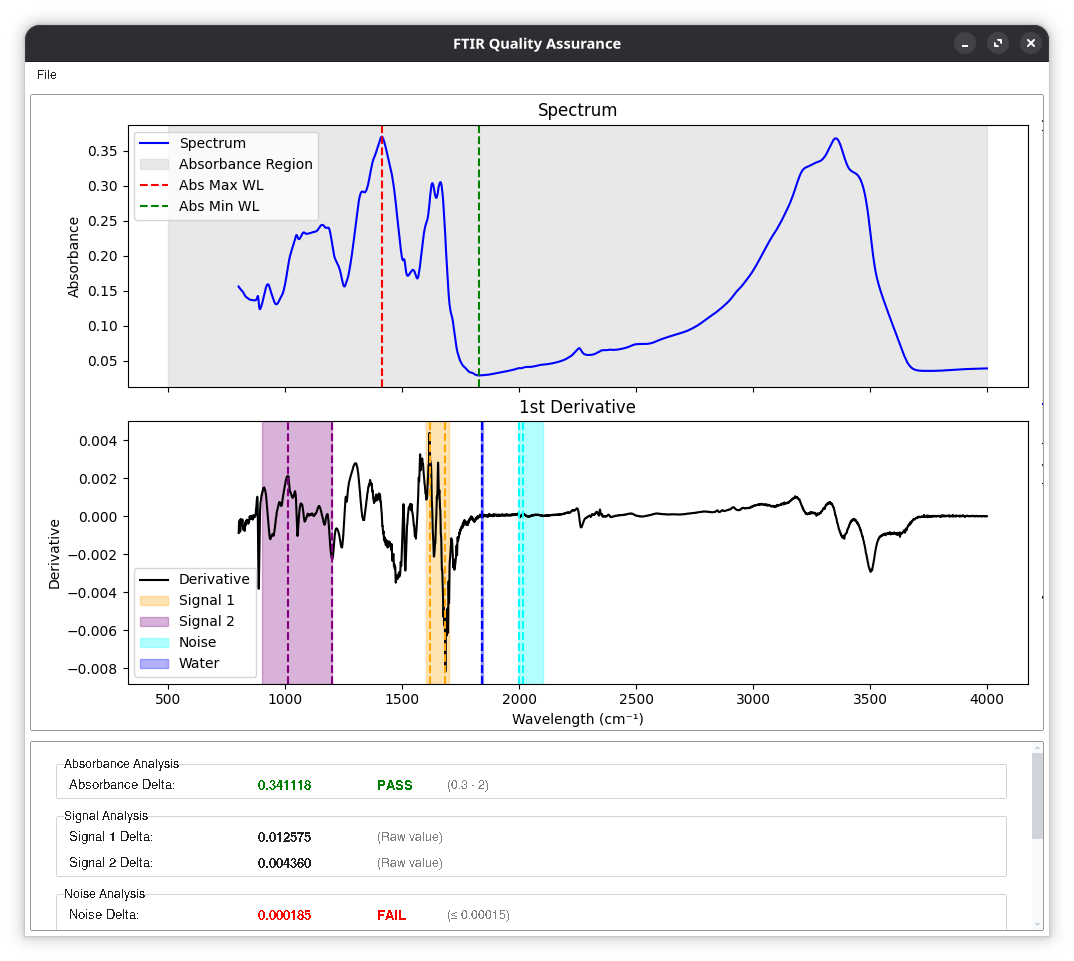
The tool is implemented as a desktop GUI in Python using Tkinter and ttkbootstrap, with NumPy, Pandas, SciPy, and Matplotlib handling data processing and visualisation. Users load spectra from CSV files, and the application automatically performs a series of domain-specific quality checks, including:
- Absorbance range checks within a configurable wavelength region, ensuring the overall dynamic range of the spectrum is appropriate.
- Signal and noise characterisation, using first-derivative spectra to estimate signal strength in key bands and noise in a dedicated region. From this, the tool computes signal-to-noise ratios (SNR) against configurable thresholds.
- Water vapour analysis, quantifying the signal in a known water vapour region and comparing it with the main signal regions to derive signal-to-water ratios. This helps detect atmospheric contamination and poor measurement conditions.
- Derivative computation, supporting both simple gradients and Savitzky–Golay filtering for more robust derivative estimation, selectable via configuration.
All analysis parameters (regions, thresholds, derivative method, theme, logging options) are configurable via a dedicated settings window and can be persisted to a configuration file. This makes the tool adaptable to different instruments, protocols, and future changes in quality criteria without needing code changes.
The application presents results in two complementary ways:
-
Visual diagnostics:
- Plots of the raw spectrum and its first derivative.
- Highlighted regions for absorbance, signals, noise, and water vapour, with markers at key maxima and minima.
- This allows users to visually inspect spectra alongside numeric metrics.
-
Structured, interpretable metrics:
- A scrollable panel summarising each test (absorbance delta, noise level, SNRs, water signal, signal-to-water ratios) with numeric values, limits, and colour-coded PASS/FAIL labels.
- An overall PASS/FAIL summary that aggregates the outcome of all checks.
- The results can be exported as a text report for record-keeping or integration into downstream processes.
From a data science perspective, the project demonstrates:
- Signal processing and feature engineering for spectroscopy data (derivatives, region-wise statistics, SNR and contamination metrics).
- Design of automated QA criteria that are robust yet interpretable to non-specialists.
- User-centred design for technical tools, turning low-level numerical checks into an interface lab technicians can use without needing to understand the underlying algorithms.
- Production-minded engineering: configuration management, logging (including optional rotating file logs), and a responsive GUI with sensible defaults.
In practice, the tool is used by lab technicians to evaluate the quality of collected samples. Because the analysis runs immediately after loading a spectrum, technicians receive instant feedback on the effect of changes in preparation and collection procedures. This has supported a more streamlined and consistent workflow for obtaining high-quality samples, reducing trial-and-error and helping to standardise best practices in the lab.